ANR (2021 - 2025) : DivAlps
Diversification and adaptation along environmental gradients : Identification of genes and traits involved in species differentiation
in alpine butterflies
Team involved : MEEBIO
Platform involved : AAEM, MEMOBIO, PASTIS
Main contact person : Laurence DESPRES (LECA, UGA)
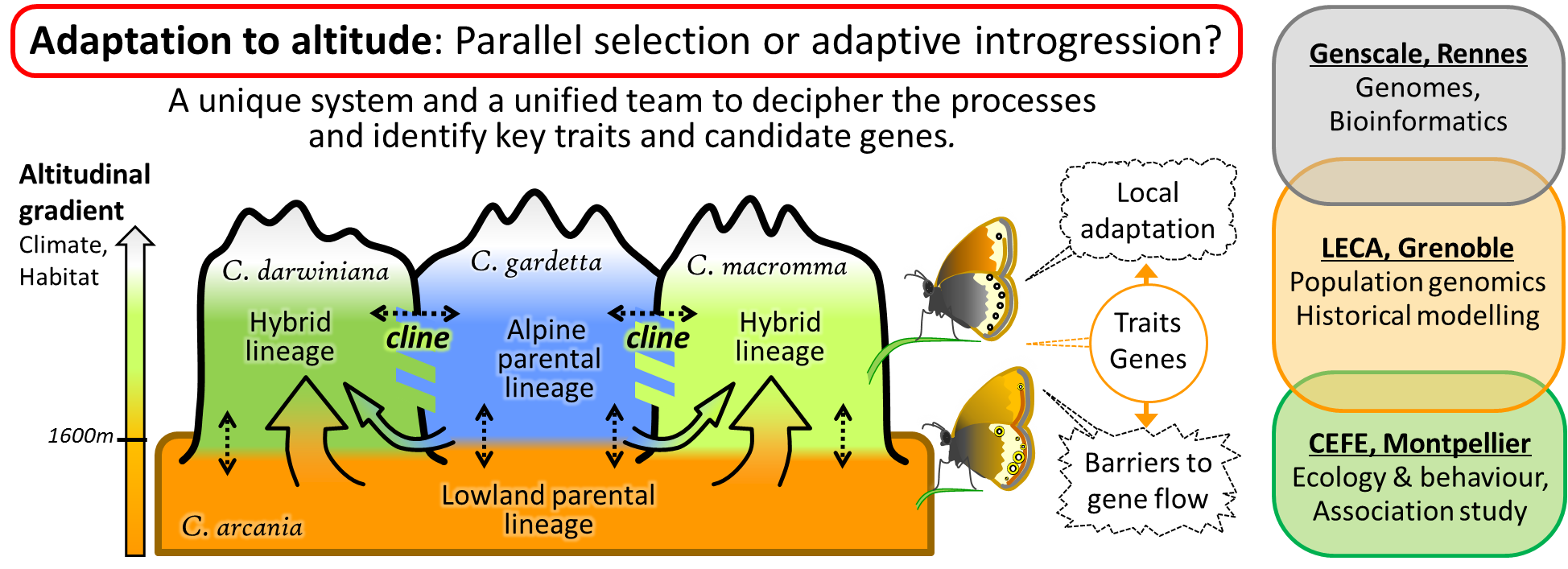
Repeated adaptation in related lineages to similar environmental conditions could result from natural selection acting independently in each lineage, or from adaptive introgression between lineages during periods of range overlap.
Here we focus on a complex of butterfly species distributed along the altitudinal gradient, and with different histories of altitudinal adaptation, to understand how populations adapt to higher elevation. We will analyse genomes of butterflies in contact zones to identify introgressions and rearrangements between taxa, i.e. regions more or less permeable to gene flow, and associate them with adaptive phenotypic variation. By using hybrid taxa originating from ancient hybridization, we will untangle the effects of genome-wide differentiation due to allopatry and demography from those of selection on genes involved in local adaptation and reproductive isolation. We will reveal to what extent these genes were exchanged between lineages. To understand how the key traits conferring altitudinal adaptation are shared by introgression among alpine lineages or act as barriers to gene flow, we will use admixed populations in contact zones, taking advantage of a natural recombination experiment allowing the segregation of the phenotypic traits characterising each taxon through many generations of recombination. This will allow linking traits and candidate genes with adaptation to changes in climatic and biotic conditions with altitude.
This system offers an excellent opportunity to decipher the processes involved in adaptation to new conditions along the altitudinal gradient, and identify the key traits and candidate genes involved. This project will mobilise forces from three labs with expertise in bioinformatics, population genomics, ecology, and experimental approaches.
Figure 1. DIVALPS graphical overview.
Major questions, study system and research consortium
Partners :
- LECA - CNRS - Université Grenoble Alpes - Université Savoie Mont Blanc (Laboratoire d’Écologie Alpine)
– Laurence DESPRES
– Thibaut CAPBLANCQ (Post-doc)
– Jesus MAVAREZ
– Matthew GREENWOOD (PhD)
– Delphine RIOUX
– Christian MIQUEL
– Frédéric LAPORTE
– Julien RENAUD
– Maya GUEGUEN
- TIMC-IMAG - CNRS - Université Grenoble Alpes (Recherche Translationnelle et Innovation en Médecine et Complexité)
– Olivier FRANCOIS
- CEFE - CNRS - Montpellier (Centre d’Ecologie Fonctionnelle et Evolutive)
– Mathieu JORON
– Anne-Geneviève BAGNERES
– Paul DONIOL-VALCROZ (PhD)
- GenScale - Inria - Université Rennes 1 (Scalable, Optimized and Parallel Algorithms for Genomics)
– Claire LEMAITRE
– Fabrice LEGEAI
– Sandra ROME (PhD)
PhD Students :
– Matthew GREENWOOD (2021-2024), Université Grenoble Alps, cosupervised by Laurence DESPRES and Thibaut CAPBLANCQ
"Genomic signals of speciation in the Coenonympha butterflies"
– Paul DONIOL-VALCROZ (2021-2024), CEFE-CNRS, Montpellier, cosupervised by Mathieu JORON and Anne-Geneviève BAGNERES
"Phenotypic variation and genomic bases of the characters involved in adaptation and diversification in alpine butterflies of the genus Coenonympha"
– Sandra ROME (2021-2024), Université Rennes 1, supervised by Claire LEMAITRE
"Structural variants in Coenonympha butterfly genomes"
Post-doc :
– Thibaut CAPBLANCQ(2021-2023)
 The federation
The federation Intranet
Intranet